Get Started
All trained models and code are publicly available. Explore the project to unlock new possibilities in protein engineering.
DMS Performance Analysis
A key challenge in protein engineering is predicting the effects of mutations. ESMS VAE excels in this domain, demonstrating superior performance on Deep Mutational Scanning (DMS) datasets from Protein Gym.
When evaluated across 162 datasets, ESMS VAE achieved a mean Spearman's ρ of 0.7779. This significantly outperforms the supervised model Kermut, which had a mean Spearman constant of 0.6982 on the same datasets. This high correlation indicates that the model's latent space effectively captures the functional impact of mutational changes.

Key Capabilities
Reconstruction & Generalization
Achieved a 97.17% reconstruction rate on a test set randomly sampled from UniRef50, proving its ability to generalize to diverse proteins.
Novel Protein Generation
Generated sequences show a maximum identity of only ~10% with training data, confirming the model creates entirely new proteins rather than copies.
Fluorescent Protein (FP) Analysis
- Classification: 98.7% 5-fold CV accuracy in classifying FPs and non-FPs.
- Regression: Low RMSE values of 2.7nm (excitation) and 3.8nm (emission) for wavelength prediction.
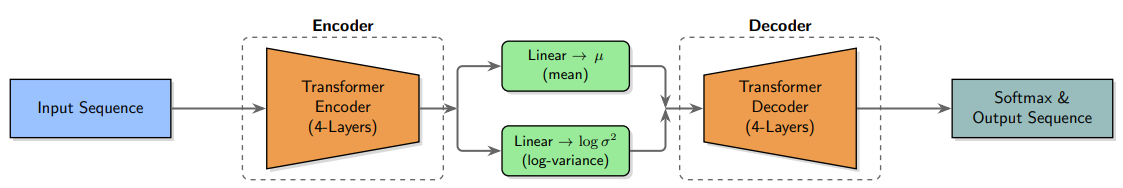
Architecture & Training
ESMS VAE is a lightweight 5.5M-parameter transformer composed of four encoder and four decoder layers. Training uses a custom structural loss based on ESMS embeddings so the latent space captures three-dimensional features. On a UniRef50 subset the model reached 97.17% reconstruction accuracy and remained robust even when noise was added.

Team & Citation
This project was developed by Danny Ahn, Shihyun Moon, Jooyoung Jung, Minjae Lee and Jeongsu Park. For full methodology and references, please refer to the project paper.
Questions? Contact ahnd6474@gmail.com.